Modern laboratory research demands precise methodologies for analyzing modified cellular components. We provide comprehensive guidance for detecting and characterizing these critical molecular modifications through established scientific protocols.
Our approach integrates three essential analytical techniques. Western blotting offers reliable detection using advanced substrate systems and specialized gel matrices. Single molecule analysis delivers unprecedented resolution for individual molecular events.
Mass spectrometry complements these methods through peptide mapping and molecular dissociation capabilities. This combination ensures accurate identification of modification sites while maintaining experimental reproducibility.
You will master fundamental protein electrophoresis techniques from sample preparation through result interpretation. Our methodology covers critical aspects including detection strategies, analytical workflows, and quality control measures. These proven approaches deliver consistent results across diverse research applications.
Key Takeaways
- Three core techniques provide comprehensive analysis: Western blotting, single molecule detection, and mass spectrometry
- Protein electrophoresis serves as the foundation for reliable molecular modification detection
- Sample preparation protocols directly impact experimental accuracy and reproducibility
- Integrated analytical workflows enhance detection sensitivity and specificity
- Quality control measures ensure consistent results across laboratory applications
- Peptide mapping techniques enable precise modification site identification
Understanding Phosphorylated Proteins
Phosphorylation serves as a fundamental regulatory mechanism that transforms protein structure and function through the strategic addition of phosphate groups. This critical post-translational modification enables cells to respond rapidly to environmental changes and internal signals. We recognize phosphorylated proteins as essential components in virtually every cellular process, from basic metabolism to complex developmental programs.
The scope of phosphorylation in biological systems is remarkable. Current research databases document over 479,000 different post-translational modifications across the human proteome. Among these modifications, more than 290,000 represent phosphorylation events occurring on serine, threonine, and tyrosine residues.

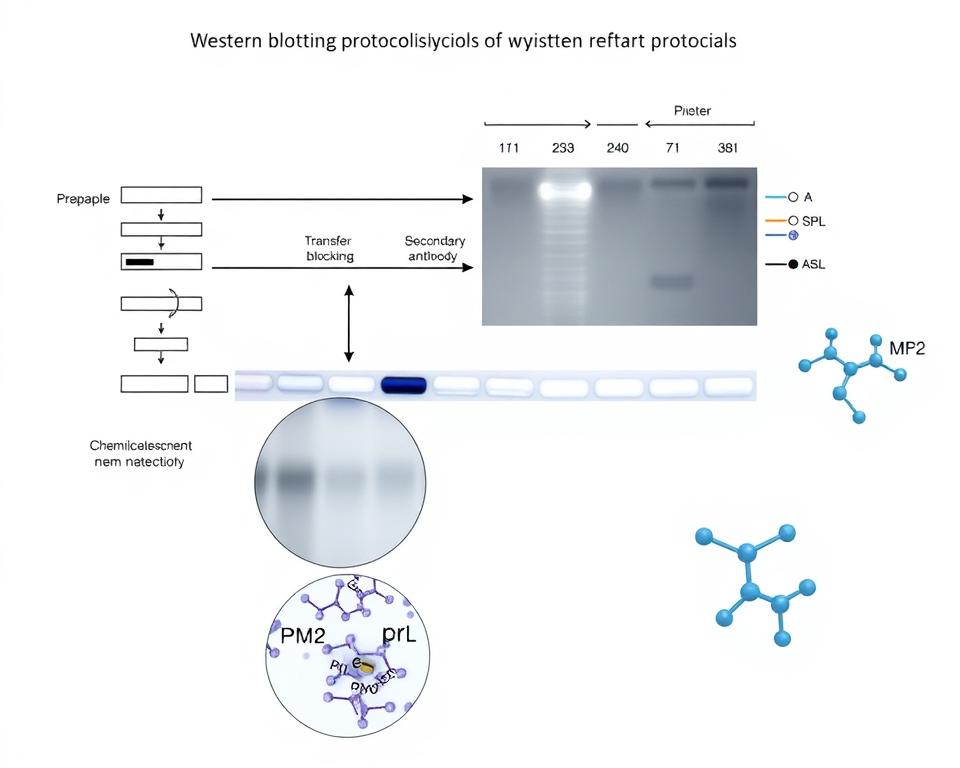
Western Blot Analysis of PI3K and Phospho – PI3K Expression
Definition of Phosphorylation
Phosphorylation involves the enzymatic addition of phosphate groups to specific amino acid residues within target proteins. This experimental protocol requires precise understanding of how kinase enzymes catalyze these reactions. The human genome encodes over 500 different kinases, each with distinct substrate specificities and regulatory mechanisms.
The process typically targets three amino acids: serine, threonine, and tyrosine. Serine and threonine phosphorylation accounts for approximately 90% of all phosphorylation events. Tyrosine phosphorylation, while less common, plays crucial roles in growth factor signaling and cellular transformation.
Each phosphorylation event creates a negative charge that can dramatically alter protein conformation. This structural change often modifies protein-protein interactions, enzymatic activity, or subcellular localization. The reversible nature of phosphorylation, mediated by phosphatase enzymes, provides cells with dynamic regulatory control.
Importance in Cellular Processes
Phosphorylated proteins regulate virtually every aspect of cellular function through coordinated signaling networks. Metabolic pathways depend heavily on phosphorylation to control enzyme activity and flux through biochemical reactions. Key regulatory enzymes like glycogen phosphorylase and acetyl-CoA carboxylase undergo phosphorylation-dependent activation or inhibition.
Cell cycle progression requires precise phosphorylation timing of cyclins and cyclin-dependent kinases. These modifications ensure proper chromosome segregation and prevent genomic instability. DNA repair mechanisms also rely on phosphorylation cascades to detect damage and coordinate repair responses.
Gene expression control involves phosphorylation of transcription factors, chromatin remodeling complexes, and RNA polymerase machinery. This allows cells to rapidly adjust protein synthesis in response to changing conditions without requiring new protein production.
Role in Signal Transduction
Signal transduction pathways utilize phosphorylation as their primary communication mechanism between cellular compartments. When you design an experimental protocol for studying these pathways, understanding phosphorylation dynamics becomes essential for accurate data interpretation.
Growth factor receptors initiate signaling through autophosphorylation events that create docking sites for downstream effector proteins. These initial phosphorylation events trigger cascading reactions that amplify signals throughout the cell. The MAP kinase pathway exemplifies this amplification, where single receptor activation can generate thousands of phosphorylated target proteins.
Stress response pathways also depend on phosphorylation networks to coordinate cellular protection mechanisms. Heat shock, oxidative stress, and nutrient deprivation all trigger specific phosphorylation patterns that activate appropriate survival responses. Understanding these patterns helps researchers develop targeted therapeutic interventions for disease states involving dysregulated signaling.

Phosphorylated-Proteins
Common Methods for Analyzing Phosphorylated Proteins
The analysis of phosphorylated proteins requires specialized detection methods tailored to specific research objectives. We present three established analytical approaches that offer distinct advantages for phosphoprotein characterization. Each technique provides unique capabilities for detecting phosphorylation events in biological samples.
Effective phosphoprotein sample preparation forms the foundation of successful analysis across all detection methods. You must consider protein stability, phosphatase inhibition, and sample integrity when selecting your analytical approach.
Western Blotting Techniques
Western blotting has served as the gold standard for phosphorylated protein identification in research laboratories. This technique utilizes phospho-specific antibodies to detect target proteins with high specificity. You can achieve reliable results through careful antibody selection and optimized blocking conditions.
The method excels in targeted protein analysis where you need to examine specific phosphorylation sites. Phosphoprotein sample preparation for Western blotting requires immediate protein extraction with phosphatase inhibitors. This approach prevents dephosphorylation during sample processing.
Western blotting provides semi-quantitative data through band intensity analysis. The technique works well for comparing phosphorylation states between different experimental conditions.
Mass Spectrometry Approaches
Mass spectrometry has emerged as the preferred method for comprehensive phosphoprotein analysis due to superior specificity. This technology enables precise localization of phosphorylation sites through advanced peptide fragmentation techniques. Modern instruments deliver unmatched analytical precision for complex protein mixtures.
Three primary dissociation methods enhance phosphopeptide identification capabilities. Collision-induced dissociation (CID) provides reliable fragmentation for most phosphopeptides. Electron transfer dissociation (ETD) preserves labile phosphate groups during analysis. Electron capture dissociation (ECD) offers complementary fragmentation patterns for comprehensive site mapping.
Phosphopeptide mapping through mass spectrometry requires specialized phosphoprotein sample preparation protocols. You must optimize digestion conditions and enrichment strategies to maximize phosphopeptide recovery.
ELISA for Detection
ELISA methods provide quantitative assessment capabilities suitable for high-throughput screening applications. This approach offers excellent reproducibility and standardization across multiple samples. You can process large sample sets efficiently using automated ELISA platforms.
The technique works particularly well for biomarker studies and clinical applications. ELISA detection requires less specialized equipment compared to mass spectrometry approaches. Phosphoprotein sample preparation for ELISA involves straightforward protein extraction and dilution procedures.
Commercial ELISA kits are available for many phosphorylated proteins of clinical interest. This availability reduces method development time and ensures consistent results across laboratories.
| Method | Specificity | Throughput | Sample Requirements | Cost Considerations |
|---|---|---|---|---|
| Western Blotting | High for targeted proteins | Low to moderate | Moderate protein amounts | Low equipment cost |
| Mass Spectrometry | Highest precision | Moderate | Minimal sample required | High equipment cost |
| ELISA Detection | Good for known targets | Very high | Small sample volumes | Moderate ongoing costs |
| Phosphoprotein Sample Preparation | Critical for all methods | Varies by protocol | Depends on technique | Method-dependent |
Our comparative analysis demonstrates that combining multiple detection approaches enhances analytical confidence. You achieve comprehensive phosphorylation characterization by selecting methods based on your specific research objectives. Each technique contributes unique strengths to the overall analytical strategy.
Protocol for Sample Preparation
We establish comprehensive sample preparation methodologies that maintain protein integrity throughout the extraction process. Sample preparation represents the most critical phase in phosphorylated protein analysis. Your success depends on implementing proper protocols that preserve phosphorylation states while eliminating interfering substances.
Effective sample preparation requires careful attention to buffer composition, temperature control, and timing. We recommend following standardized protocols that have proven successful in research laboratories worldwide. The following procedures ensure optimal results for downstream analysis.
Cell Lysis Techniques
Cell lysis forms the foundation of protein extraction from cellular samples. We utilize the SiMPull protocol, which employs 1% IGEPAL CA-630 in 50 mM Tris pH 7.2/150 mM NaCl buffer system. This detergent concentration provides optimal membrane disruption without denaturing target proteins.
Protease and phosphatase inhibitors must be added immediately before lysis. These inhibitors prevent protein degradation and maintain phosphorylation states during extraction. We recommend preparing fresh inhibitor cocktails for each experiment to ensure maximum effectiveness.
Temperature control during lysis prevents protein denaturation. Keep samples on ice throughout the procedure. Gentle agitation helps achieve complete cell disruption without generating excessive heat through mechanical stress.
Protein Extraction Methods
Protein extraction methods vary depending on your target proteins and sample types. We implement standardized buffer systems that maintain protein stability while facilitating efficient extraction. The Bio-Rad protocol includes specific buffer preparations optimized for phosphorylated protein analysis.
TBST Wash Buffer preparation involves combining 1x TBS with 0.1% Tween 20. This buffer system provides optimal washing conditions for subsequent purification steps. Casein Tween Blocking Buffer serves as an effective blocking agent in immunodetection applications.
Electrophoresis buffer composition directly impacts protein separation quality during gel electrophoresis. We formulate buffers with precise pH control and ionic strength optimization. These parameters ensure consistent protein migration patterns and reliable results.
| Buffer Component | Concentration | Function | Storage Conditions |
|---|---|---|---|
| IGEPAL CA-630 | 1% | Cell membrane disruption | Room temperature |
| Tris pH 7.2 | 50 mM | pH buffering system | 4°C storage |
| NaCl | 150 mM | Ionic strength maintenance | Room temperature |
| Tween 20 | 0.1% | Protein solubilization | Room temperature |
Precipitation and Cleanup Steps
Precipitation techniques concentrate proteins while removing interfering substances from your samples. We employ selective precipitation methods that preserve protein structure and phosphorylation states. These steps eliminate salts, detergents, and other contaminants that interfere with analysis.
Pervanadate treatment at 1 mM concentration serves as a positive control for phosphorylation studies. This treatment inhibits protein tyrosine phosphatases and maintains phosphorylation levels during sample processing. We recommend including pervanadate controls in every experiment.
Cleanup procedures remove excess salts and buffer components that affect electrophoresis buffer performance. Proper cleanup ensures optimal protein migration and reduces background interference. For comprehensive guidance on optimizing these procedures, refer to our western blot technical guidelines for detailed protocols.
Final sample preparation involves protein concentration determination and dilution to appropriate working concentrations. We recommend using Bradford or BCA assays for accurate protein quantification. Store prepared samples at -80°C for long-term stability or proceed immediately to electrophoresis.
Running SDS-PAGE for Phosphorylated Proteins
SDS-PAGE electrophoresis serves as the foundation for accurate phosphorylated protein analysis in laboratory settings. We recommend following specific experimental protocol guidelines to achieve optimal separation and resolution of your protein samples. The success of downstream detection methods depends entirely on proper gel preparation and electrophoresis conditions.
Phosphorylated proteins require careful handling during protein electrophoresis to maintain their structural integrity. You must consider molecular weight variations caused by phosphate group additions when selecting appropriate gel concentrations and running parameters.
Gel Preparation Guidelines
We utilize Bio-Rad Criterion 4-15% TGX Stain-Free gels for optimal phosphorylated protein separation. These gradient gels provide excellent resolution across a wide molecular weight range, accommodating proteins from 10 kDa to 250 kDa effectively.
The gradient concentration ensures proper separation of closely related protein species. You should verify gel integrity before use by checking for cracks or air bubbles that could affect migration patterns.
Store prepared gels at 4°C and use within the manufacturer’s recommended timeframe. Temperature control during storage prevents gel degradation and maintains consistent performance.
Sample Loading Techniques
Precise sample loading requires careful attention to volume control and lane organization. We recommend loading samples immediately after gel preparation to prevent diffusion effects that compromise band sharpness.
Use the following loading specifications for optimal results:
- Load 10-30 μg of total protein per well
- Maintain consistent sample volumes across all lanes
- Include molecular weight standards in designated lanes
- Avoid overfilling wells to prevent sample spillover
Mix Precision Plus All Blue Standards and Precision Plus Unstained Standards at a 1:1 ratio. Load exactly 10 μL per well of this marker mixture to ensure accurate molecular weight determination.
Electrophoresis Conditions
We run gels using 1x TGS Running Buffer at 300V constant voltage. This experimental protocol typically requires 20-25 minutes for complete separation, depending on gel size and protein composition.
Monitor the dye front progression throughout the run. Stop electrophoresis when the dye front reaches the bottom of the gel to prevent protein loss and maintain optimal resolution.
The following table outlines critical electrophoresis parameters for phosphorylated protein electrophoresis:
| Parameter | Setting | Duration | Expected Result |
|---|---|---|---|
| Voltage | 300V constant | 20-25 minutes | Sharp band separation |
| Current | Auto-regulated | Throughout run | Consistent migration |
| Temperature | 4°C cooling | Entire procedure | Reduced band distortion |
| Buffer Volume | 1x TGS, 1L total | Single use | Optimal conductivity |
Temperature control during electrophoresis prevents heat-induced protein denaturation. We recommend using a cooling system to maintain 4°C throughout the run, especially for temperature-sensitive phosphorylated proteins.
Buffer quality significantly impacts separation efficiency. Replace running buffer between experiments to maintain consistent ionic strength and pH levels. Fresh buffer ensures reproducible migration patterns and prevents artifacts that could interfere with phosphorylation analysis.
Document gel images immediately after electrophoresis completion. This practice allows you to assess separation quality before proceeding to transfer steps in your experimental protocol.
Transfer Techniques for Western Blots
The transfer process represents a critical step in maintaining phosphorylation integrity during Western blot procedures. We must carefully select appropriate transfer methods to ensure complete protein migration while preserving phosphorylated states. Phosphoprotein sample preparation requires specific considerations during membrane transfer to maintain protein functionality.
You can achieve optimal results by maintaining cold temperatures throughout the transfer process. The buffer pH must remain stable to prevent protein degradation. Our methodology focuses on preserving phosphorylation sites during the entire transfer procedure.
Options for Protein Transfer
We offer three primary transfer methods for phosphoprotein sample preparation. Wet transfer provides the most complete protein migration for high molecular weight proteins. This method uses fully submerged gel and membrane assemblies in transfer buffer.
Semi-dry transfer offers faster processing times with reduced buffer volumes. You can complete transfers in 30-60 minutes depending on protein size. Rapid transfer systems provide the quickest option for routine phosphoprotein analysis.
The Bio-Rad Trans-Blot Turbo RTA Mini/Midi Transfer Kit represents our recommended rapid transfer solution. This system uses low fluorescence PVDF membranes for stain-free imaging capabilities. UV activation enables total protein normalization for accurate quantification.
Optimizing Transfer Efficiency
Transfer efficiency depends on several critical factors during phosphoprotein sample preparation. Membrane selection directly impacts protein binding capacity and background signal levels. PVDF membranes offer superior protein retention compared to nitrocellulose options.
You must consider protein molecular weight when selecting transfer conditions. High molecular weight proteins require extended transfer times or higher voltage settings. Transfer duration affects both efficiency and protein integrity preservation.
Temperature control prevents protein degradation during the transfer process. We recommend maintaining temperatures below 4°C for phosphoprotein applications. Buffer composition must support both protein migration and phosphorylation state maintenance.
Transfer Buffer Recipes
Standard transfer buffer formulations require modification for phosphoprotein sample preparation. We recommend Tris-glycine buffer systems with methanol concentrations between 10-20%. Lower methanol percentages improve high molecular weight protein transfer efficiency.
Bis-Tris buffer systems provide enhanced pH stability during extended transfers. You can add protease inhibitors directly to transfer buffers for additional protein protection. Buffer pH should remain between 8.0-8.5 for optimal transfer conditions.
| Buffer Type | Composition | Transfer Time | Best Application |
|---|---|---|---|
| Standard Tris-Glycine | 25mM Tris, 192mM Glycine, 20% Methanol | 60-120 minutes | General phosphoproteins |
| Low Methanol | 25mM Tris, 192mM Glycine, 10% Methanol | 90-180 minutes | High molecular weight |
| Bis-Tris System | 25mM Bis-Tris, 25mM Bicine, 1mM EDTA | 45-90 minutes | pH-sensitive proteins |
| Rapid Transfer | 48mM Tris, 39mM Glycine, 20% Methanol | 7-15 minutes | Routine analysis |
Our transfer protocols ensure complete protein migration while maintaining phosphorylation integrity. You achieve consistent results by following these established buffer formulations and transfer conditions. Phosphoprotein sample preparation success depends on careful attention to these transfer methodology details.
Detection Methods for Phosphorylated Proteins
Detection methods for phosphorylated proteins form the cornerstone of successful experimental protocol development. We recommend establishing a systematic approach that balances sensitivity requirements with practical laboratory constraints. Your choice of detection methodology directly impacts the accuracy and reliability of phosphorylation analysis results.
The selection process involves evaluating multiple factors including target protein abundance, required sensitivity levels, and downstream analysis needs. Proper method selection ensures optimal signal-to-noise ratios while minimizing non-specific binding interference.
Antibody Selection Criteria
Selecting appropriate antibodies requires careful evaluation of several critical parameters. We establish specific criteria to ensure reliable detection and minimize experimental variability in your experimental protocol.
- Specificity validation: Verify antibody recognition of target phosphorylation sites without cross-reactivity
- Sensitivity assessment: Determine minimum detection limits for your protein concentration range
- Reproducibility testing: Confirm consistent results across multiple experimental batches
- Species compatibility: Ensure antibody recognition across relevant biological systems
- Application suitability: Match antibody performance to intended detection methods
Quality antibodies demonstrate consistent performance across different experimental conditions. We recommend validating each antibody lot before implementing in critical experiments.
Using Phospho-Specific Antibodies
Phospho-specific antibodies provide targeted detection of specific phosphorylation events. The Bio-Rad protocol utilizes Phospho-Specific PrecisionAb Antibodies at 1:1,000 dilution in Casein Tween Blocking Buffer for optimal results.
Overnight incubation at 4°C ensures complete antibody binding while maintaining protein integrity. This extended incubation period allows thorough penetration and specific binding to target phosphorylation sites.
SiMPull methodology employs AF647-conjugated anti-phosphotyrosine antibodies for direct labeling approaches. This technique eliminates secondary antibody steps while providing excellent signal clarity.
We recommend including appropriate controls to validate antibody specificity. Positive and negative controls help distinguish specific phosphorylation signals from background noise.
Chemiluminescence vs. Colorimetric Detection
Detection method selection significantly impacts experimental sensitivity and quantitative accuracy. We compare both approaches to help you choose the optimal detection system.
Chemiluminescence detection offers superior sensitivity for low-abundance phosphorylated proteins. The Clarity Western ECL Substrate Kit provides reliable chemiluminescent detection with 1:1 component mixing ratios.
This method enables detection of femtogram quantities while maintaining excellent signal-to-noise ratios. Enhanced sensitivity proves particularly valuable when analyzing subtle phosphorylation changes.
Colorimetric detection provides direct visual results without specialized imaging equipment. This approach offers good reproducibility for high-abundance targets but shows limited sensitivity compared to chemiluminescent methods.
We recommend chemiluminescence for research applications requiring maximum sensitivity. Colorimetric methods work well for routine quality control and educational demonstrations where moderate sensitivity suffices.
Interpretation of Results
Understanding your experimental data begins with careful evaluation of band intensity and phosphorylation patterns. We establish systematic approaches that ensure accurate interpretation of phosphorylated protein analysis results. Proper data analysis requires attention to multiple factors including loading controls, transfer efficiency, and detection sensitivity variations.
Successful result interpretation depends on implementing appropriate normalization controls and statistical methods. You must consider experimental variables that affect data quality and reproducibility. Electrophoresis buffer consistency plays a crucial role in maintaining reliable comparative results between different experimental conditions.
Analyzing Band Intensity
Band intensity analysis forms the foundation of quantitative phosphorylated protein assessment. We recommend using densitometric measurements to evaluate signal strength accurately. ImageLab software provides reliable tools for measuring band intensities with consistent parameters across all samples.
Proper background subtraction ensures accurate intensity calculations. You should establish baseline measurements from areas adjacent to protein bands. Consistent imaging conditions prevent variations that could affect quantitative comparisons.
Normalization against loading controls eliminates variations from unequal protein loading. We use housekeeping proteins like β-actin or GAPDH as reference standards. This approach ensures that observed differences reflect actual phosphorylation changes rather than loading inconsistencies.
Comparing Phosphorylation States
Effective phosphorylation state comparison requires systematic experimental design with appropriate controls. Bio-Rad protocol includes mock and phosphatase-treated samples for validation purposes. Lambda Protein Phosphatase treatment serves as a negative control to confirm phospho-specific antibody binding.
We establish baseline phosphorylation levels using untreated control samples. Treatment conditions should be compared against these baseline measurements. Statistical significance testing validates observed differences between experimental groups.
Phosphatase treatment controls demonstrate antibody specificity for phosphorylated forms. You can verify that signal reduction occurs after phosphatase treatment. This validation step confirms that detected bands represent genuine phosphorylation events.
Quantitative Analysis Techniques
Advanced quantitative methods provide precise measurements of phosphorylation levels. SiMPull methodology offers single molecule quantification with optimal receptor density of 0.04-0.08 receptors per μm². This technique enables detection of individual phosphorylation events.
Ratio calculations compare phosphorylated protein levels to total protein amounts. We calculate phospho-to-total protein ratios for accurate assessment. These ratios eliminate variations from total protein expression changes.
Statistical validation ensures reliable interpretation of quantitative results. You should perform multiple independent experiments for statistical power. Standard deviation calculations and significance testing provide confidence in observed differences.
- Densitometric analysis – Quantifies band intensity using specialized software
- Ratio calculations – Compares phosphorylated to total protein levels
- Statistical testing – Validates significance of observed differences
- Control validation – Confirms antibody specificity through phosphatase treatment
Troubleshooting Common Issues
We encounter various technical obstacles when analyzing phosphorylated proteins that demand specific solutions. These challenges can compromise your experimental results and require systematic approaches to identify root causes. Understanding common problems helps you maintain consistent data quality throughout your research.
Successful troubleshooting involves examining each step of your protocol systematically. From sample preparation through detection, multiple factors can influence your final results. We provide practical solutions to address the most frequent issues researchers face.
Band Smearing and Lack of Signal
Band smearing typically results from improper gel preparation or inadequate sample handling during protein electrophoresis. You can resolve this issue by ensuring complete polymerization of your gel matrix before loading samples. Allow sufficient time for gel setting and maintain consistent temperature conditions.
Lack of signal often stems from insufficient protein loading or degraded phosphorylation states. Check your protein concentration using Bradford or BCA assays before electrophoresis. Phosphoprotein sample preparation requires immediate processing with phosphatase inhibitors to prevent dephosphorylation.
Optimize your electrophoresis conditions by reducing voltage and extending run times. High voltage generates excessive heat, causing band distortion and poor resolution. Use fresh running buffer and ensure proper electrode connections throughout the process.
Non-Specific Binding Problems
Non-specific binding creates false positive signals that compromise data interpretation. Systematic antibody validation helps eliminate cross-reactivity issues with related proteins. Test different antibody concentrations to establish optimal working dilutions for your specific samples.
Blocking optimization prevents unwanted protein interactions on membrane surfaces. Use appropriate blocking agents such as BSA or non-fat milk for 1-2 hours at room temperature. Extend blocking time for problematic antibodies or increase blocking agent concentration.
Refine your washing protocols using TBST buffer with proper pH and salt concentrations. The Bio-Rad protocol emphasizes thorough washing procedures to prevent chamber-to-chamber contamination. Increase wash frequency and duration for persistent background issues.
Overlapping Bands in Gel
Overlapping bands occur when proteins with similar molecular weights migrate together during protein electrophoresis. Gradient gel selection provides better separation resolution compared to uniform percentage gels. Choose appropriate gradient ranges based on your target protein sizes.
Sample concentration adjustment improves band definition and reduces overcrowding effects. Dilute concentrated samples or reduce loading volumes to prevent lane overloading. Phosphoprotein sample preparation should maintain consistent protein amounts across all samples.
Optimize separation conditions by extending electrophoresis time or reducing sample volumes. Lower acrylamide concentrations help separate larger proteins, while higher concentrations improve small protein resolution. Consider using bis-tris gels for enhanced separation of phosphorylated protein variants.
Applications of Phosphorylated Protein Analysis
Modern experimental protocols for phosphorylated protein analysis enable breakthrough discoveries in disease mechanisms and drug development. These methodologies have transformed our understanding of cellular signaling networks. We can now identify therapeutic targets with unprecedented precision.
The versatility of phosphorylated protein analysis extends across multiple research domains. From basic science investigations to clinical diagnostics, these techniques provide valuable insights. You can apply these methods to address complex biological questions and medical challenges.
Disease Research, Including Cancer
Cancer research represents one of the most significant applications of phosphorylated protein analysis. Over 180 kinases are implicated in various diseases, with cancer showing the highest prevalence of kinase dysregulation. These enzymes control critical cellular processes including growth, division, and death.
The pharmaceutical industry has responded with remarkable success. More than 60 kinase inhibitors have received clinical approval for cancer treatment. Additionally, over 150 kinase inhibitors are currently advancing through clinical trials.
We utilize experimental protocols to identify specific phosphorylation patterns in tumor samples. This approach reveals which signaling pathways drive cancer progression. You can then target these pathways with precision therapies.
- Tyrosine kinase inhibitors for leukemia treatment
- Serine/threonine kinase targeting in solid tumors
- Combination therapies based on phosphorylation profiles
- Resistance mechanism identification through phospho-analysis
Pathway Mapping and Drug Discovery
Pathway mapping through phosphorylated protein analysis reveals complex regulatory networks. Cardiac myosin binding protein-C (cMyBP-C) regulation demonstrates this complexity. Multiple kinases including PKA, PKCε, and RSK2 target different phosphorylation sites on this single protein.
Each kinase creates distinct functional outcomes. PKA phosphorylation enhances cardiac contractility. PKCε modification affects protein stability. RSK2 targeting influences protein-protein interactions.
Drug discovery teams leverage these experimental protocols to understand mechanism of action. We can determine how potential therapeutics alter phosphorylation patterns. This knowledge guides lead compound optimization and safety assessment.
- Target identification through phospho-proteomics
- Mechanism elucidation via phosphorylation mapping
- Efficacy assessment using phospho-specific readouts
- Safety evaluation through off-target phosphorylation analysis
Biomarker Identification in Clinical Settings
Clinical diagnostics increasingly rely on phosphorylated protein biomarkers. These markers provide superior specificity compared to total protein measurements. Phosphorylation states reflect real-time cellular activity rather than just protein abundance.
Disease diagnosis benefits from phosphorylation pattern analysis. We can distinguish between different disease subtypes based on kinase activity profiles. This precision enables personalized treatment selection.
Treatment monitoring represents another critical application. You can track therapeutic response by measuring changes in target protein phosphorylation. This approach provides early indicators of treatment efficacy or resistance development.
| Clinical Application | Phospho-Biomarker | Disease Context | Clinical Utility |
|---|---|---|---|
| Cardiac injury | Phospho-troponin | Myocardial infarction | Early diagnosis |
| Cancer prognosis | Phospho-p53 | Various cancers | Treatment selection |
| Neurodegeneration | Phospho-tau | Alzheimer’s disease | Disease staging |
| Diabetes monitoring | Phospho-insulin receptor | Type 2 diabetes | Therapy optimization |
The integration of phosphorylated protein analysis into clinical workflows continues expanding. These experimental protocols bridge the gap between research discoveries and patient care. We anticipate further growth in phospho-biomarker applications across diverse medical specialties.
Future Trends in Phosphorylated Protein Research
The landscape of phosphorylated protein analysis continues evolving rapidly. We observe transformative developments that will reshape how you approach protein research in coming years. These advances promise enhanced precision and broader clinical applications.
Advances in Detection Technologies
Single molecule techniques like SiMPull now enable quantification of phosphorylation heterogeneity at individual protein levels. Mass spectrometry sensitivity improvements allow detection of previously undetectable phosphorylation events. Real-time monitoring systems provide dynamic insights into cellular signaling cascades. Electrophoresis buffer formulations continue advancing through automated optimization systems that enhance reproducibility across laboratories.
Integration with Systems Biology
Phosphoproteomics integration with systems biology approaches reveals comprehensive cellular signaling networks. You can now map complex regulatory relationships and understand dynamic signaling responses. Artificial intelligence assists in data interpretation, identifying patterns that traditional analysis methods miss. These integrated approaches provide deeper understanding of protein function within biological systems.
Implications for Personalized Medicine
Phosphorylation-based biomarker development opens new pathways for precision medicine. We anticipate targeted therapy selection based on individual phosphorylation profiles. Treatment response prediction becomes more accurate through phosphoproteomic analysis. Multiplexed detection platforms will enable simultaneous analysis of multiple phosphorylation states, streamlining clinical diagnostics and research workflows for improved patient outcomes.
References and further readings:
1.Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22(10):545-554.
https://linkinghub.elsevier.com/retrieve/pii/S01689525060026302.Engholm-Keller K, Larsen MR. Technologies and challenges in large-scale phosphoproteomics. Proteomics. 2013;13(6):910-931.
https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pmic.2012004843.Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9(6):1451-1468.
https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pmic.2008004544.Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94-101.
https://www.nature.com/articles/nbt1046
FAQ
What is phosphorylation and why is it important in cellular processes?
We define phosphorylation as the critical post-translational modification that involves adding phosphate groups to serine, threonine, and tyrosine residues. This modification fundamentally alters protein structure and activity, serving as a molecular switch that controls numerous physiological processes from gene expression to metabolic regulation. Phosphorylation creates functional diversity within the proteome, enabling complex cellular responses through coordinated signaling cascades.
Which analytical methods are most effective for detecting phosphorylated proteins?
We present three established analytical methods, each offering distinct advantages. Western blotting provides reliable detection through phospho-specific antibodies for targeted protein analysis. Mass spectrometry delivers comprehensive identification capabilities with precise phosphorylation site localization through peptide fragmentation analysis. ELISA detection offers quantitative assessment suitable for high-throughput screening applications. Our comparative analysis demonstrates that combining multiple detection approaches enhances analytical confidence.
What are the critical requirements for phosphoprotein sample preparation?
We establish that proper phosphoprotein sample preparation requires implementing appropriate cell lysis techniques using specific detergent concentrations and buffer systems to maintain protein integrity. You must include phosphatase inhibitors during protein extraction to prevent dephosphorylation during processing. Precipitation and cleanup steps eliminate interfering substances while concentrating target proteins. Maintaining sample integrity through controlled temperature conditions and optimized electrophoresis buffer formulations ensures reliable detection of phosphorylated protein species.
How should I optimize SDS-PAGE conditions for phosphorylated protein analysis?
We provide detailed guidelines emphasizing that you must prepare gradient gels according to specific concentration requirements for optimal separation across molecular weight ranges. Protein electrophoresis parameters including voltage, current, and run time must be carefully controlled to achieve consistent separation quality. Sample loading requires precise volume control and proper lane organization. Our experimental protocol emphasizes using molecular weight standards for accurate protein identification and phosphorylation state assessment.
What transfer techniques work best for Western blot analysis of phosphorylated proteins?
We detail that you can choose between wet, semi-dry, and rapid transfer systems based on experimental requirements. Optimizing transfer efficiency requires careful consideration of membrane type, electrophoresis buffer composition, and transfer duration to ensure complete protein migration. Transfer buffer recipes must maintain protein integrity while facilitating efficient membrane binding. Our methodology ensures optimal protein transfer while preserving phosphorylation states throughout the process.
How do I select appropriate antibodies for phosphorylated protein detection?
We establish that you must consider specificity, sensitivity, and cross-reactivity profiles when selecting antibodies for target phosphorylation sites. Using phospho-specific antibodies requires careful validation and optimization to ensure reliable detection without non-specific binding. Chemiluminescence detection offers superior sensitivity compared to colorimetric methods, enabling detection of low-abundance phosphorylated proteins. Our experimental protocol includes detailed procedures for antibody incubation and signal development optimization.
What is the best approach for interpreting phosphorylated protein analysis results?
We establish systematic approaches requiring you to analyze band intensity variations using appropriate normalization controls and statistical methods for accurate quantification. Comparing phosphorylation states requires careful consideration of loading controls, transfer efficiency, and detection sensitivity variations. Quantitative analysis techniques include densitometric measurements, ratio calculations, and statistical validation. Electrophoresis buffer consistency directly impacts result reproducibility and comparative accuracy between experimental conditions.
How can I troubleshoot common issues like band smearing and non-specific binding?
We address that you can resolve band smearing through optimized gel preparation, proper sample handling, and controlled electrophoresis conditions. Non-specific binding problems require systematic antibody validation, blocking optimization, and washing protocol refinement. Overlapping bands necessitate gradient gel selection and sample concentration adjustment. Protein electrophoresis troubleshooting involves systematic evaluation of buffer composition, gel integrity, and transfer efficiency. Phosphoprotein sample preparation issues often stem from inadequate phosphatase inhibition or improper storage conditions.
What are the main applications of phosphorylated protein analysis in research?
We demonstrate diverse applications including disease research, particularly cancer studies where kinase dysregulation drives pathological processes. Pathway mapping applications reveal complex signaling networks controlling cellular responses. Drug discovery efforts benefit through target identification, mechanism elucidation, and therapeutic efficacy assessment. Biomarker identification in clinical settings enables disease diagnosis, prognosis determination, and treatment monitoring through phosphorylation pattern analysis.
What future trends are emerging in phosphorylated protein research?
We examine significant advances in detection technologies including enhanced mass spectrometry sensitivity, single molecule analysis capabilities, and real-time phosphorylation monitoring systems. Integration with systems biology approaches enables comprehensive network analysis revealing complex regulatory relationships. Implications for personalized medicine include phosphorylation-based biomarker development and targeted therapy selection. Future developments will focus on multiplexed detection platforms, artificial intelligence-assisted data interpretation, and clinical translation of phosphoproteomics discoveries.
Leo Bios
Hello, I’m Leo Bios. As an assistant lecturer, I teach cellular and
molecular biology to undergraduates at a regional US Midwest university. I started as a research tech in
a biotech startup over a decade ago, working on molecular diagnostic tools. This practical experience
fuels my teaching and writing, keeping me engaged in biology’s evolution.





Leave a Comment
Your email address will not be published. Required fields are marked *